Journal of Primeasia
Targeting p38 MAPK: Molecular Docking and Therapeutic Insights for Alzheimer’s Disease Management
Md Abu Bakar Siddique1*, Asim Debnath2, Md Sefaut Ullah3, Md Sakil Amin4, Azizur Rahman5, Moushumi Afroza Mou1, Md Abdur Rahman Biswash6, Md Shaikh Bin Noor Tamim7, Mst. Shahana Akter8, Bulbul Ahmed9, Abdullah Al Numan9, Md Mahedi Hasan Shabuj9
Journal of Primeasia 6 (1) 1-11 https://doi.org/10.25163/primeasia.6110116
Submitted: 31 October 2024 Revised: 03 January 2025 Published: 04 January 2025
Abstract
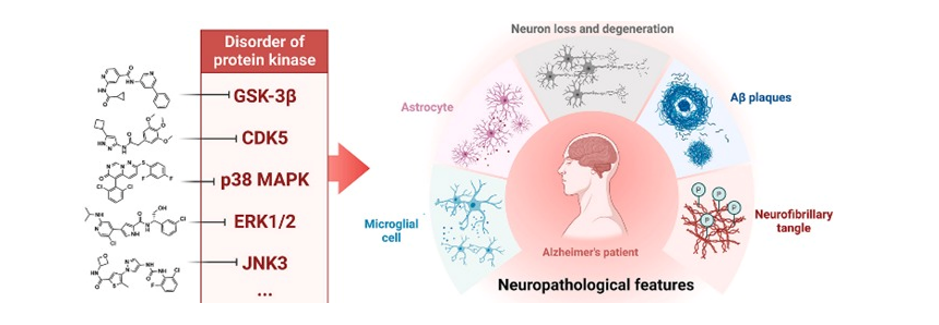
Background: Alzheimer’s disease (AD), a progressive neurodegenerative disorder, is closely linked to the p38 mitogen-activated protein kinase (MAPK) pathway, which regulates neuroinflammation, oxidative stress, apoptosis, and other key pathological processes. Developing selective inhibitors targeting p38 MAPK could offer novel therapeutic interventions for AD. Methods: Nine molecules, selected for their therapeutic potential based on literature, were docked against p38 MAPK (5MTX) using molecular docking. Binding affinities, hydrogen bonding, and hydrophobic interactions were analyzed to assess the strength and stability of ligand-receptor interactions. Residues contributing to selectivity and therapeutic potential were identified, and results were contextualized with in vitro and in vivo studies. Results: NJK14047 exhibited the highest binding affinity (-10.2 kcal/mol) due to hydrogen bonds with Asn115, Gly110, Met109, Asp168, and Glu71, contributing to enhanced stability and selectivity. Ginsenoside Rg1 (-7.9 kcal/mol) and Apigenin (-8.7 kcal/mol) demonstrated significant interactions with key residues, including Thr106 and Leu104, with Ginsenoside Rg1 supporting mitophagy and memory improvement in AD models. Skepinone-L showed high inhibitory activity with hydrophobic residues but lacked hydrogen bonding. In vivo studies supported the neuroprotective and anti-inflammatory effects of several candidates, with NJK14047 reducing microglial activation and promoting neuroprotection. Conclusion: The study underscores the therapeutic potential of p38 MAPK inhibitors in AD management. NJK14047, with its strong binding affinity and selectivity, emerges as a lead candidate for further exploration. These findings highlight the need for clinical trials to validate the efficacy of p38 MAPK inhibitors as a comprehensive approach to treating AD.
Keywords: Alzheimer’s Disease (AD), p38 MAPK Inhibitors, Molecular Docking, Neuroinflammation, Therapeutic Targets
References
Ashwell, J. D. (2006). The many paths to p38 mitogen- activated protein kinase activation in the immune system. 6(July), 532–540. https://doi.org/10.1038/nri1865
Avital, A., Goshen, I., Kamsler, A., Segal, M., Iverfeldt, K., Richter-Levin, G., & Yirmiya, R. (2003). Impaired interleukin-1 signaling is associated with deficits in hippocampal memory processes and neural plasticity. Hippocampus, 13(7), 826–834. https://doi.org/10.1002/hipo.10135
Bains, J. S., & Oliet, S. H. R. (2007). Glia: they make your memories stick! Trends in Neurosciences, 30(8), 417–424. https://doi.org/10.1016/j.tins.2007.06.007
Barbieri, R., Alloisio, S., Ferroni, S., & Nobile, M. (2008). Differential crosstalk between P2X7 and arachidonic acid in activation of mitogen-activated protein kinases. Neurochemistry International, 53(6–8), 255–262. https://doi.org/10.1016/j.neuint.2008.05.001
Barone, F. C., Irving, E. A., Ray, A. M., Lee, J. C., Kassis, S., Kumar, S., Badger, A. M., White, R. F., McVey, M. J., Legos, J. J., Erhardt, J. A., Nelson, A. H., Ohlstein, E. H., Hunter, A. J., Ward, K., Smith, B. R., Adams, J. L., & Parsons, A. A. (2001). SB 239063, a second-generation p38 mitogen-activated protein kinase inhibitor, reduces brain injury and neurological deficits in cerebral focal ischemia. Journal of Pharmacology and Experimental Therapeutics, 296(2), 312–321.
Bernardo, A., & Minghetti, L. (2007). PPAR-γ Agonists as Regulators of Microglial Activation and Brain Inflammation. Current Pharmaceutical Design, 12(1), 93–109. https://doi.org/10.2174/138161206780574579
Bhat, N. R., Feinstein, D. L., Shen, Q., & Bhat, A. N. (2002). p38 MAPK-mediated transcriptional activation of inducible nitric-oxide synthase in glial cells: Roles of nuclear factors, nuclear factor κB, cAMP response element-binding protein, CCAAT/enhancer-binding protein-β, and activating transcription factor-2. Journal of Biological Chemistry, 277(33), 29584–29592. https://doi.org/10.1074/jbc.M204994200
Bhat, N. R., Zhang, P., Lee, J. C., & Hogan, E. L. (1998). Extracellular signal-regulated kinase and p38 subgroups of mitogen- activated protein kinases regulate inducible nitric oxide synthase and tumor necrosis factor-α gene expression in endotoxin-stimulated primary glial cultures. 18(5), 1633–1641.
Bliss, T. V. P., & Collingridge, G. L. (1993). A synaptic model of memory: Long-term potentiation in the hippocampus. Nature, 361(6407), 31–39. https://doi.org/10.1038/361031a0
Hodges, J. (2017). Pick’s disease: Its relationship to progressive aphasia, semantic dementia and frontotemporal dementia. In Dementia, Fifth Edition (Vol. 21). https://doi.org/10.1201/9781315381572
Knutson, B., Westdorp, A., Kaiser, E., Hommer, D., Beattie, E. C., Stellwagen, D., Morishita, W., Bresnahan, J. C., Ha, B. K., & Zastrow, M. Von. (2002). <Beattie Science 2002.pdf>. 295(March), 2282–2285.
Welch, L., Lewitter, F., Schwartz, R., Brooksbank, C., Radivojac, P., Gaeta, B., & Schneider, M. V. (2014). Bioinformatics curriculum guidelines: Toward a definition of core competencies. Computational Biology, 10(3). https://doi.org/10.1371/journal.pcbi.1003496
Xu, Y., Yan, J., Zhou, P., Li, J., Gao, H., Xia, Y., & Wang, Q. (2012). Neurotransmitter receptors and cognitive dysfunction in Alzheimer’s disease and Parkinson’s disease. Progress in Neurobiology, 97(1), 1–13. https://doi.org/10.1016/j.pneurobio.2012.02.002
Yan, Z., Zhou, Z., Wu, Q., Chen, Z. B., Koo, E. H., & Zhong, S. (2020). Presymptomatic increase of an extracellular RNA in blood plasma associates with the development of Alzheimer’s disease. Current Biology, 30(10), 1771-1782.e3. https://doi.org/10.1016/j.cub.2020.02.084
Yang, C., Zhu, Z., Tong, B. C. K., Iyaswamy, A., Xie, W. J., Zhu, Y., Sreenivasmurthy, S. G., Senthilkumar, K., Cheung, K. H., Song, J. X., Zhang, H. J., & Li, M. (2020). A stress response p38 MAP kinase inhibitor SB202190 promoted TFEB/TFE3-dependent autophagy and lysosomal biogenesis independent of p38. Redox Biology, 32. https://doi.org/10.1016/j.redox.2020.101445
Yang, S., Zhou, G., Liu, H., Zhang, B., Li, J., Cui, R., & Du, Y. (2013). Protective effects of p38 MAPK inhibitor SB202190 against hippocampal apoptosis and spatial learning and memory deficits in a rat model of vascular dementia. BioMed Research International, 2013. https://doi.org/10.1155/2013/215798
Yang, T. T., Liu, C. G., Gao, S. C., Zhang, Y., & Wang, P. C. (2018). The serum exosome derived microRNA-135a, -193b, and -384 were potential Alzheimer’s disease biomarkers. Biomedical and Environmental Sciences, 31(2), 87–96. https://doi.org/10.3967/bes2018.011
Yasuda, S., Sugiura, H., Tanaka, H., Takigami, S., & Yamagata, K. (2011). p38 MAP kinase inhibitors as potential therapeutic drugs for neural diseases. Central Nervous System Agents in Medicinal Chemistry, 11.
Yokota, T., & Wang, Y. (2016). P38 MAP kinases in the heart. Gene, 575(2), 369–376. https://doi.org/10.1016/j.gene.2015.09.030
Yuan, H., Ma, Q., Ye, L., & Piao, G. (2016). The traditional medicine and modern medicine from natural products. Molecules, 21(5). https://doi.org/10.3390/molecules21050559
Yue, Q., Song, Y., Liu, Z., Zhang, L., Yang, L., & Li, J. (2022). Receptor for advanced glycation end products (RAGE): A pivotal hub in immune diseases. Molecules, 27(15). https://doi.org/10.3390/molecules27154922
Yusufzai, S. K., Khan, M. S., Sulaiman, O., Osman, H., & Lamjin, D. N. (2018). Molecular docking studies of coumarin hybrids as potential acetylcholinesterase, butyrylcholinesterase, monoamine oxidase A/B, and β-amyloid inhibitors for Alzheimer’s disease. Chemistry Central Journal, 12(1). https://doi.org/10.1186/s13065-018-0497-z
Zarubin, T., & Han, J. (2005). Activation and signaling of the p38 MAP kinase pathway. Cell Research, 15(1).
Zetterberg, H., Skillbäck, T., Mattsson, N., Trojanowski, J. Q., Portelius, E., Shaw, L. M., Weiner, M. W., & Blennow, K. (2016). Association of cerebrospinal fluid neurofilament light concentration with Alzheimer disease progression. JAMA Neurology, 73(1), 60–67. https://doi.org/10.1001/jamaneurol.2015.3037
Zhang, F., & Jiang, L. (2014). Neuroinflammation in Alzheimer’s disease. Neuropsychiatric Disease and Treatment, 11, 243–256. https://doi.org/10.2147/NDT.S75546
Zhang, N., Zhang, L., Li, Y., Gordon, M. L., Cai, L., Wang, Y., Xing, M., & Cheng, Y. (2017). Urine AD7c-NTP predicts amyloid deposition and symptom of agitation in patients with Alzheimer’s disease and mild cognitive impairment. Journal of Alzheimer’s Disease, 60(1), 87–95. https://doi.org/10.3233/JAD-170383
Zhang, R., Zhu, X., Bai, H., & Ning, K. (2019). Network pharmacology databases for traditional Chinese medicine: Review and assessment. Frontiers in Pharmacology, 10(February). https://doi.org/10.3389/fphar.2019.00123
Zhang, W. (2016). Network pharmacology: A further description. Network Pharmacology, 1(1).
Zhang, Y., Yang, S., Jiao, Y., Liu, H., Yuan, H., Lu, S., Ran, T., Yao, S., Ke, Z., Xu, J., Xiong, X., Chen, Y., & Lu, T. (2013). An integrated virtual screening approach for VEGFR-2 inhibitors. Journal of Chemical Information and Modeling, 53(12), 3163–3177. https://doi.org/10.1021/ci400429g
Zhao, L., Wang, J. L., Liu, R., Li, X. X., Li, J. F., & Zhang, L. (2013). Neuroprotective, anti-amyloidogenic and neurotrophic effects of apigenin in an Alzheimer’s disease mouse model. Molecules, 18(8), 9949–9965. https://doi.org/10.3390/molecules18089949
Zheng, S., Xue, T., Wang, B., Guo, H., & Liu, Q. (2022). Application of network pharmacology in the study of mechanism of Chinese medicine in the treatment of ulcerative colitis: A review. Frontiers in Bioinformatics, 2. https://doi.org/10.3389/fbinf.2022.928116
Recommended articles
Network Pharmacology and Docking-Based Evaluation of Quercetin as an Inhibitor of PI3K Pathway in Glioblastoma
Integrating Clinical Data and Molecular Profiling to Predict Antibiotic-Induced Anaphylaxis: A Comparative Study of Ceftriaxone and Meropenem
TGF-β1 as a Molecular Target and Vitamin D Interaction in Diabetic Nephropathy: A Bio-clinical Correlation Study
Save
Citation
View
Share
